Athenomics - Immune repertoire analysis by NGS
Next-generation sequencing (NGS) has revolutionized immune repertoire analysis, enabling researchers to profile millions of TCR/BCR sequences in a single experiment
Introduction to Immune Repertoire Sequencing
The immune system’s ability to recognize and combat pathogens depends on the extraordinary diversity of T-cell receptors (TCRs) and B-cell receptors (BCRs). These receptors are generated through V(D)J recombination, a highly variable genetic process that produces a vast array of unique immune cells, collectively known as the immune repertoire. At any moment, an individual’s immune repertoire is composed of approximately 10^8 lymphocytes, each with distinct specificities.
Accurately profiling the immune repertoire is a complex challenge. It requires high-throughput methods to capture the enormous number of genetic rearrangements present, as well as high sensitivity to identify rare receptor clones. Next-generation sequencing (NGS) has fundamentally transformed immune repertoire analysis, allowing researchers to:
- Profile millions of TCR and BCR sequences in a single experiment
- Track clonal expansion in conditions such as cancer, autoimmunity, and infections
- Discover biomarkers for predicting immunotherapy responses
Understanding TCR and BCR Diversity
T-Cell Receptors (TCRs):
Found on T cells, TCRs recognize peptide-MHC complexes. TCRs are composed of either α/β or γ/δ chains, with most studies focusing on the TCRβ chain. The receptor diversity is generated via V(D)J recombination, especially affecting the hypervariable complementarity-determining region 3 (CDR3).
B-Cell Receptors (BCRs):
Expressed by B cells, BCRs recognize antigens directly. BCRs consist of heavy (IgH) and light (Igκ/Igλ) chains. Their diversity is generated through V(D)J recombination during B cell development, followed by somatic hypermutation (SHM) and affinity maturation during immune responses. This process allows for an immense theoretical repertoire—up to 10^{25} distinct receptor combinations.
The sequencing and analysis of TCR and BCR repertoires have significant applications in cancer research, infectious disease monitoring, vaccine development, and the diagnosis and treatment of immune-related diseases.
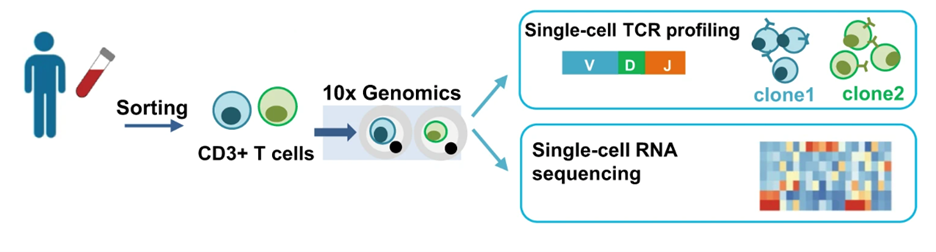
We have expertise in characterizing TCR and BCR repertoires using 10x single-cell sequencing technology. If you need a customized T/BCR sequencing solution, please contact us. We can help you design an experiment tailored to your specific research needs, free of charge.
Source of figure: DOI: 10.1038/s41467-022-29175-x