Athenomics - Introduction to Single-Cell RNA-seq
scRNA-seq measures the distribution of expression levels for each gene at single-cell resolution, enabling researchers to study cellular heterogeneity in unprecedented detail.
RNA-seq is widely applied to samples containing mixed cell populations, a method often referred to as bulk RNA-seq. This approach serves diverse purposes, such as characterizing gene expression across tissues in healthy versus diseased states, comparing wild-type and mutant conditions, or differentiating between control and treated samples. RNA-seq also plays a critical role in evolutionary research by enabling comparative transcriptomic analyses of tissues sampled from different species. Beyond quantifying known transcripts, RNA-seq can uncover and annotate novel genes, alternative isoforms, and other previously uncharacterized transcripts in both model and non-model organisms. However, bulk RNA-seq only yields an averaged gene expression profile across every cell in a sample, thereby masking the underlying heterogeneity among individual cells. As a consequence, this approach is inadequate for investigating highly variable biological systems, such as tissues in early development or complex organs like the brain, where cell-to-cell differences are fundamental.
To overcome this limitation, single-cell RNA sequencing (scRNA-seq) was developed, with the first successful application published in 2009. The technology became widespread around 2014 as refinements in protocols and decreasing sequencing costs made it broadly accessible. Unlike bulk RNA-seq, which reflects an average gene expression across populations, scRNA-seq measures the distribution of expression levels for each gene at single-cell resolution, enabling researchers to study cellular heterogeneity in unprecedented detail. This capability has unlocked new insights in biological research, including the identification of novel or rare cell types, detection of compositional shifts between healthy and disease tissues, and characterization of dynamic differentiation trajectories during development. One of the most transformative applications of scRNA-seq is the construction of gene atlases, which map cellular diversity across entire organisms and serve as foundational resources for biomedical and basic science research.
Modern scRNA-seq studies routinely profile thousands to millions of cells, and study sizes continue to increase as the technology advances. Researchers can select from a variety of protocols, both commercial and open-source, each offering different trade-offs in sensitivity, throughput, cost, and technical requirements. There is currently a wide diversity of protocols for generating scRNA-seq data, each with unique strengths and weaknesses.
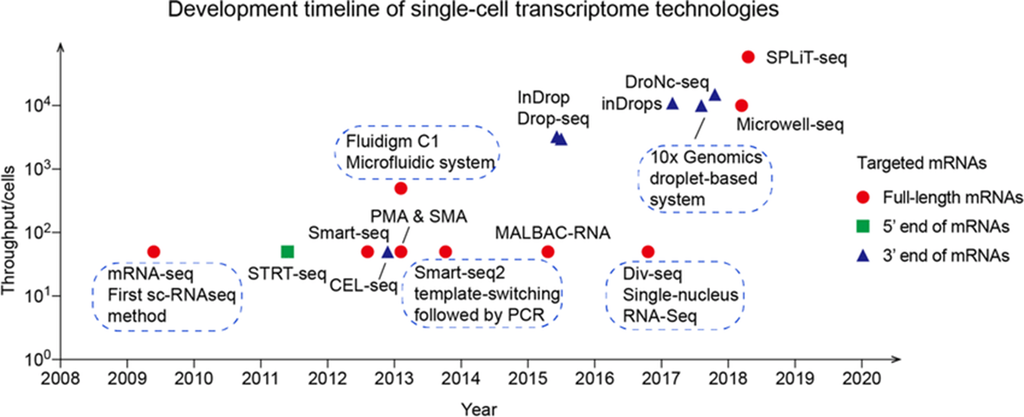
What Protocol Should I Choose?
Designing a single-cell RNA-seq experiment requires careful consideration of multiple technical and biological factors. Important practical decisions include cost per cell, the desired number of cells, and sequencing depth, all of which directly influence the protocol selection. Equally important are experimental design considerations, such as managing batch effects that may arise from processing samples at different times, as well as ensuring adequate biological replication to support downstream analysis and address research questions robustly.
The platform choice should be informed by the specific biological objectives of the study. For comprehensive profiling of heterogeneous tissues, droplet-based protocols are ideal thanks to their high throughput and ability to minimize bias in cell capture. Alternatively, when working with well-defined populations distinguished by known cell markers, fluorescence-activated cell sorting (FACS) followed by deep sequencing of selected cells often yields superior results.
Full-length transcript sequencing is essential for studies focused on analyzing alternative splicing and isoform diversity, as tagged methods (5’ and 3’) capture only transcript ends and provide limited information about isoform structure. In contrast, unique molecular identifiers (UMIs), which are only compatible with tagged protocols, enhance gene-level quantification accuracy. When studying rare cell populations without established markers, researchers generally need to sequence a larger total number of cells to capture enough examples of these populations. While this increases costs, it remains the most reliable method for detecting rare, low-abundance cell types.
We currently offer tagged (5’ and 3’) scRNA-seq services, which significantly reduce library preparation and sequencing costs. Please reach out if you are planning to analyze gene expression at the individual cell level—this approach provides insights into cellular heterogeneity, rare cell populations, and the mechanisms of disease.
Source of figure: DOI: 10.1007/s12015-022-10330-2