Athenomics - Microbiome Analysis using Next-Generation Sequencing (NGS)
Metagenomics provides researchers with unprecedented insights into microbial diversity, function, and dynamics across various environments - from the human gut to extreme ecosystems
Next-generation sequencing (NGS) has transformed microbiome research by enabling comprehensive and quantitative analysis of microbial communities. Unlike traditional culture-based methods, metagenomic sequencing can identify both culturable and unculturable microorganisms, as well as detect low-abundance species that would otherwise go unnoticed. This approach provides unprecedented insights into microbial diversity, function, and dynamics across various environments, from the human gut to extreme ecosystems.
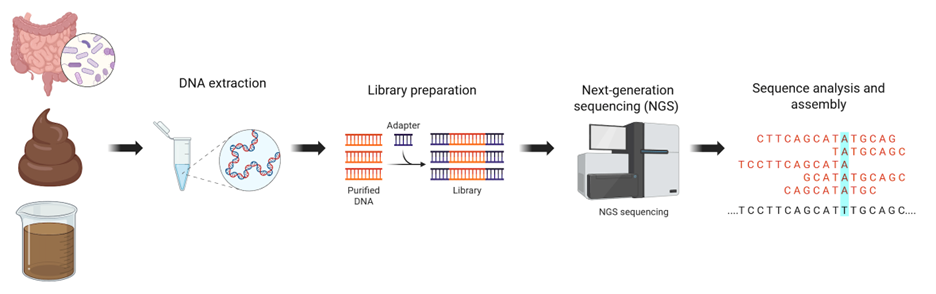
However, successful microbiome studies depend on robust experimental design and careful execution. Key aspects—such as sample collection, DNA extraction, library preparation, sequencing depth, and bioinformatic analysis—must be optimized, as each plays a major role in the reliability and interpretation of results. Sequencing coverage and throughput are particularly critical to ensure sufficient data for comprehensive genetic characterization of the microbiome.
The microbiome represents a highly complex ecosystem, presenting significant challenges for data analysis. The vast volume of sequence data from thousands of microbial species and strains demands powerful bioinformatics and considerable computational resources. Complexity further increases when analyzing multiple samples from different body sites within an individual, across populations, or over time. Therefore, computational requirements and bioinformatic strategies must be considered when designing microbiome studies.
Shotgun Metagenomic Sequencing
Shotgun sequencing randomly samples and sequences DNA fragments from an entire sample, following DNA shearing and next-generation sequencing. Unlike targeted approaches, shotgun metagenomic sequencing enables comprehensive, unbiased profiling of all microbial genomes in a sample—including those from unculturable or previously unknown species. High sequencing depth allows for detection of rare and low-abundance microbial species.
Despite its power, shotgun sequencing can miss certain microbial members due to issues like contaminant DNA or the inherent size and complexity of microbiomes. Moreover, shotgun metagenomics is resource-intensive both financially and computationally. Reconstructing microbial communities from fragmented DNA requires sophisticated computational assembly techniques and relies on extensive reference databases—currently comprising over 50,000 microbial genomes—which may introduce bias. Strain-level resolution is also limited compared to targeted sequencing (such as single-isolate genome sequencing) due to the lower genomic resolution of the untargeted data.
Targeted Metagenomic Sequencing
Targeted sequencing focuses on specific genes or genomic regions of interest, leveraging established genomic knowledge to enhance sequencing coverage. This strategy streamlines downstream data analysis and interpretation and reduces overall workflow complexity and cost relative to untargeted methods.
16S Ribosomal RNA (rRNA) Gene Sequencing
16S rRNA gene sequencing is the gold standard for bacterial taxonomic profiling and general microbial community analysis. Its value lies in the structure of the 16S rRNA gene, which contains both highly conserved regions (enabling broad phylogenetic comparisons) and nine hypervariable segments (V1–V9) that serve as species-specific identification markers. These variable regions, part of the small ribosomal subunit, allow accurate bacterial and archaeal identification while maintaining evolutionary context.
Earlier 16S analyses often used PCR amplification of just one or a few variable regions, giving limited resolution. Advances in NGS technology now enable simultaneous examination of multiple V regions, vastly improving discriminatory power. This enhanced capacity is especially critical for clinical applications, where comprehensive sequencing of V regions improves detection and identification of pathogenic bacteria.
High-throughput NGS not only enhances V region sequencing but also allows for multiplexed, advanced analyses. Targeted methods can now distinguish closely related bacterial species and enable sensitive detection of antimicrobial resistance genes. As our understanding of the interplay between microbiomes and hosts grows, targeted sequencing is proving invaluable for biomarker discovery, disease association studies, and for predicting therapeutic response—unlocking new opportunities in precision medicine.
Our collaborators possess comprehensive platforms to accelerate microbial discovery. Using our validated shotgun, 16S/18S/ITS sequencing and analysis capabilities, we can characterize and isolate microbes quickly and efficiently, providing cost-effective solutions for microbiome research.