Athenomics - RNA Sequencing (RNA-Seq) Methods for NGS
RNA sequencing (RNA-Seq) has revolutionized transcriptomics by enabling researchers to analyze gene expression, discover novel transcripts, and detect RNA modifications with high precision.
Introduction to RNA-Seq
RNA sequencing (RNA-Seq) has revolutionized transcriptomics by enabling researchers to analyze gene expression, discover novel transcripts, and detect RNA modifications with high precision. As a next-generation sequencing (NGS) application, RNA-Seq provides a high-resolution, genome-wide view of the transcriptome, offering significant advantages over older microarray-based approaches. Multiple RNA-Seq techniques are now available, from whole transcriptome sequencing to targeted RNA sequencing, each with specific advantages and disadvantages depending on research goals.
General RNA Sequencing Methods
There are several main approaches to RNA sequencing:
Total RNA Whole Transcriptome:
Whole transcriptome analysis examines both coding and noncoding RNA at once, making it suitable for novel discovery. However, this approach requires higher sequencing throughput to achieve appropriate coverage and can suffer from inefficiencies or bias due to diverse transcript lengths.
mRNA Sequencing:
By using poly(A) selection, mRNA sequencing targets all messenger RNA for gene expression analysis and is capable of identifying both novel and known transcripts.
Small RNA Sequencing:
This method isolates small RNA species, focusing on noncoding RNA such as microRNA (miRNA) for both novel and known content discoveries.
Targeted RNA Sequencing:
Targeted methods sequence only specific transcripts of interest. This approach is more cost-effective for the analysis of targeted genes and can be applied to many sample types, including degraded samples such as FFPE (formalin-fixed, paraffin-embedded) tissues.
General Workflow of RNA Sequencing
RNA samples are fragmented to a desirable size range to suit the read length limitations of current NGS platforms. Fragmentation can be achieved with divalent cation solutions, enzymatic approaches, or by fragmenting full-length cDNA (depending on library preparation protocols). The general workflow for RNA sequencing includes:
A. RNA Fragmentation
Mechanical (sonication) or enzymatic (using RNA fragmentation reagents) methods are used to produce fragments in the range of approximately 200–500 base pairs.
B. cDNA Synthesis
Reverse transcription converts RNA fragments to cDNA. Strand-specific protocols often employ dUTP marking.
C. Adapter Ligation & Indexing
Sequencing adapters compatible with platforms like Illumina or Nanopore are added, and unique barcodes are used for multiplexing samples.
D. PCR Amplification (Optional)
This step enhances library yield, though it may introduce amplification bias.
Choosing mRNA Sequencing Methods
mRNA sequencing is one of the most widely adopted RNA-Seq approaches. Common strategies include:
Full-Length mRNA Sequencing:
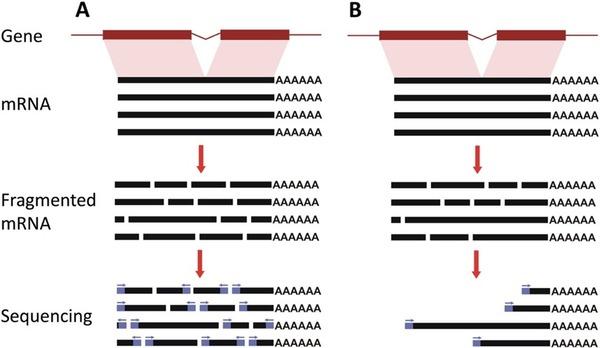
This provides comprehensive information about mRNA structure, including splicing variants and SNPs. However, a known limitation is that longer transcripts are broken into more fragments during preparation, causing statistical bias—longer transcripts end up represented by more reads than shorter ones.
3’ mRNA Sequencing:
Developed to minimize length-related bias, this method avoids fragmenting mRNAs prior to reverse transcription. Instead, each cDNA is generated from the 3′ end of the mRNA, and only one cDNA molecule per transcript is sequenced. Therefore, read counts directly reflect transcript abundance, and several studies confirm this method yields results comparable to full-length sequencing when identifying differentially expressed genes.
5’ mRNA Sequencing:
Similar to the 3’ method, 5’ mRNA sequencing enriches for the 5’ end. This is especially valuable for gene expression profiling and for identifying TCR and BCR sequences, supporting immune cell characterization and antibody screening.
Full-length mRNA-Seq typically requires approximately six times more sequencing reads per library than 3’ mRNA sequencing to achieve equivalent transcriptome coverage. The 3’ approach is easier to scale and is well suited for high-throughput, cost-effective differential gene expression studies.
Our team offers expertise in full-length, 3’, and 5’ mRNA sequencing, and we can help you select the optimal approach for your project, balancing both time and cost. Please feel free to contact us if your work involves bulk RNA-seq, single-cell RNA-seq, or V(D)J sequencing projects.
Source of figure: DOI: 10.1016/j.gdata.2016.11.002